Welcome to Fly News! I’m DinK, bringing you live updates from the notorious Alphabet City, nestled in the Left (Fly) Wing of the Science Center. This vibrant city comprises three distinct areas known as DIPstricts: A, B, and C. The residents of these DIPstricts are renowned for their inexhaustible liveliness, as demonstrated by their impressive 42 minutes and 50 seconds record of silence. While their cheerful nature is a source of amusement, it has also sparked concerns among inhabitants of neighboring lands such as The Solis and Eshelman regions. What exactly happens in this peculiar city, and what exciting activities take place within each DIPstrict to ignite such boundless enthusiasm? Today, we have mayors from each DIPstrict of Alphabet City who will provide us with an insider’s perspective on the captivating activities unfolding behind the scenes.
DIPstrict A
Greetings! I’m Y (not the abbreviation, pronounced ME without the M), and the mayor of the DIPstrict A. Dipeptidase A (DIP-A) in Drosophila melanogaster underwent the name changing process and is now proudly known as carnosine dipeptidase A, all because our human relative carnosinase called us. Well, even enzymes can have family drama.
In the human carnosinase district, carnosinemia patients face challenges due to low carnosinase activity, such as developmental delays, neurological deficits, and even seizures. On the flip side, our Drosophila residents with DIP-A deficiency struggle to survive the larval stage. Life can be tough for our tiny residents.
Talk about family resemblance! DIP-A and carnosinase share 76% similarity in their amino acid sequences. They are both dimers, made up of two subunits. The Dip-A gene produces two different versions of proteins, known as isoform A and isoform C. So, DIP-A can exist as either a homodimer (where the subunits are the same, AA and CC) or as a heterodimer (mixing it up with different subunits, AC).
To observe the presence of DIP-A and its active structures, I conducted a non-denaturing gel electrophoresis or native PAGE. This technique allows the separation of proteins in the sample without breaking them down into subunits. In combination with the Western blotting technique, where anti-DIP-A antibodies specifically detect and bind to the DIP-A protein, I aimed to identify the bands representing the presence of each protein. You’ll easily find me and the residents of DIPstrict A buzzing in the cold room since native PAGE requires low temperature. Due to the close molecular weights of the dimeric proteins, instead of neatly separated bands, residents love sticking together and end up with a big smear. I am still trying to find a better way to separate them for a good photoshoot.
I also stained the gel for DIP-A dimeric enzyme activity using a DIP-A specific substrate with an agar overlay on the gel to amplify the signal of the bands. The data I got so far from this staining technique is a smiling face (on the gel, not me). The gel is cheering me on. Who knows what other surprises await me in the cold room?

DIPstrict B
Hey! I’m Mayor Van Dinh (not to be mistaken by DinK) and welcome to DIPstrict B, a vibrant and bustling neighborhood within the genomic metropolis! DIPstrict B, aka the “Splice Street,” is where all the action happens, with dipeptidase B (DIP-B) stealing the spotlight. In the heart of DIPstrict B, we encounter the charismatic Drosophila Dip-B gene. It’s the lifeblood of this neighborhood, producing multiple mRNA isoforms that come in different flavors. We have Isoform A, Isoform B, Isoform C, Isoform D and Isoforms A/C and E, who hide intriguing secrets beneath their varying 5′ UTR lengths. They are the colorful characters shaping the genetic landscape of this district! Just as a neighborhood’s streets have diverse architectural styles, these mRNA isoforms showcase unique splicing patterns. The translated regions remain consistent, much like the beautifully designed buildings, while the 5′ UTR acts as the entrance, with each isoform having its own distinct length.

To shed light on this intriguing genetic neighborhood, residents are embarking on comparative studies to understand the relative proportions of these Dip-B mRNA isoforms. They are using RT-qPCR to quantitatively measure the mRNA isoform levels. This investigation will provide insight into how alternative splicing patterns in the 5′ UTR region influence gene expression, much like unraveling the secrets behind hidden passages in a neighborhood. Just as a neighborhood evolves over time, gene expression levels can fluctuate during different developmental stages.
By analyzing the occurrence and proportion of mRNA isoforms in various growth phases of D. melanogaster, residents can witness the genetic neighborhood transform. It’s like observing a neighborhood’s architecture adapting to the changing tastes and needs of its residents.

Understanding the intricate relationship between splicing patterns within the 5′ UTR region and transcription levels of mRNA isoforms opens a gateway to comprehend the significance of alternative gene expression. These findings could illuminate molecular mechanisms and mutations, aiding in the identification of potential therapeutic targets for human diseases such as Alzhiemers and Crohn’s. Just like how understanding a neighborhood’s dynamics helps create a better living environment, deciphering gene expression intricacies can contribute to improved health outcomes.
As you bid farewell to the lively streets of DIPstrict B, I hope you were able to witness the marvels of alternative mRNA isoforms and their impact on gene expression. This metaphorical journey through a genetic neighborhood has shown us that exploring the world of dipeptidase genes is not just about molecules and DNA but a captivating exploration of hidden stories and secrets within. As you go along your journey of discovering the great activities within each DIPstrict within Alphabet City, I hope that we can continue to unveil the wonders within DIPstrict B and unlock the mysteries of gene expression, one splice at a time!
—

DIPstrict C
DIPstrict C runs a Xaa-Pro treatment plant where dipeptidase C (DIP-C, a prolidase) in Drosophila melanogaster breaks down the bonds between the two amino acids. It is a relative of the PEPD (protein PEPD) in the human town, who runs a similar plant. The PEPD plant is essential in maintaining the overall fitness of the human, specifically protein metabolism, collagen turnover, and extracellular matrix remodeling, and hence involved in wound healing, inflammation, angiogenesis, cell proliferation, and carcinogenesis. If PEPD went on strike, humans would suffer from dysmorphia, developmental delays, and impaired wound healing. However, overtime working at the plant is observed in Alzheimer’s disease and associated with several types of cancer such as bladder cancer, breast cancer, and pancreatic cancer.
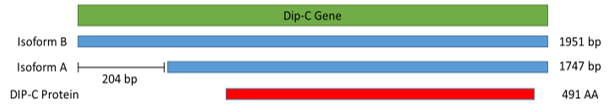
In DIPstrict C, there are two structures (mRNA isoforms), called A and B. The two are almost identical except that mRNA isoform A is 204 bp shorter than mRNA isoform B at 5’ UTR. Regardless, they are both owned by one guy (protein) called DIP-C.
Previous residents have found that structure A is more common than structure B. Since Drosophila melanogaster is a metamorphic organism, their genetics can differ greatly across developmental stages. This led me to wonder if the proportion of structures vary at different developmental stages. Is A or B more common, or are they equal in number? To find out more about this, I, Mayor Vy, use RT-qPCR (reverse transcriptase quantitative PCR), a technique that allows me to quickly and accurately quantify the proportion of the structures in the region. Total RNA from samples have been collected, RT-qPCR has been conducted, and analysis is under way.

Visitors wonder about the underlying reason for the disproportion of the two structures. Therefore, I’m also doing bioinformatic sequence analysis of the structures. Our two major suggestions are: (1) structure (mRNA isoform) A is more beautiful (better promoter) and (2) structure A is more stable.
For (1), I have been searching through the Drosophila Housing Beauty Standards aka Drosophila Promoter Database that reported features that were reported as attractive in housing from 205 samples (205 promoters). None of them match exactly. This actually might not be surprising, since the database surveyed only 205 structures (genes) out of more than 13,000 of the Drosophila genome. As for (2), I am still evaluating blueprints on how to effectively and easily examine the stability of the structures.
—
The information above was compiled by the Mayor of DIPstrict C, Vy. Vy loves DIP-C, cats, pandas, flies not flying, sleeping at midday not midnight, and eating two dinners a day. If you want to see more works from Vy, please donate lots of love, RNAse-free tips and a new comfortable hammock.
As more activities are sure to develop within these DIPstricts, Fly News will be the first to report to you with more live updates. Reporter DinK signing off.